Proteins are the workhorses that hold our cells operating, and there are numerous hundreds of kinds of proteins in our cells, every performing a specialised perform. Researchers have lengthy recognized that the construction of a protein determines what it could actually do. Extra just lately, researchers are coming to understand {that a} protein’s localization can be vital for its perform. Cells are stuffed with compartments that assist to prepare their many denizens. Together with the well-known organelles that adorn the pages of biology textbooks, these areas additionally embrace a wide range of dynamic, membrane-less compartments that focus sure molecules collectively to carry out shared capabilities. Figuring out the place a given protein localizes, and who it co-localizes with, can subsequently be helpful for higher understanding that protein and its position within the wholesome or diseased cell, however researchers have lacked a scientific method to predict this info.
In the meantime, protein construction has been studied for over half-a-century, culminating within the synthetic intelligence software AlphaFold, which might predict protein construction from a protein’s amino acid code, the linear string of constructing blocks inside it that folds to create its construction. AlphaFold and fashions prefer it have turn out to be extensively used instruments in analysis.
Proteins additionally comprise areas of amino acids that don’t fold into a hard and fast construction, however are as a substitute essential for serving to proteins be a part of dynamic compartments within the cell. MIT Professor Richard Younger and colleagues puzzled whether or not the code in these areas could possibly be used to foretell protein localization in the identical manner that different areas are used to foretell construction. Different researchers have found some protein sequences that code for protein localization, and a few have begun growing predictive fashions for protein localization. Nevertheless, researchers didn’t know whether or not a protein’s localization to any dynamic compartment could possibly be predicted primarily based on its sequence, nor did they’ve a comparable software to AlphaFold for predicting localization.
Now, Younger, additionally member of the Whitehead Institute for Organic Analysis; Younger lab postdoc Henry Kilgore; Regina Barzilay, the College of Engineering Distinguished Professor for AI and Well being in MIT’s Division of Electrical Engineering and Laptop Science and principal investigator within the Laptop Science and Synthetic Intelligence Laboratory (CSAIL); and colleagues have constructed such a mannequin, which they name ProtGPS. In a paper printed on Feb. 6 within the journal Science, with first authors Kilgore and Barzilay lab graduate college students Itamar Chinn, Peter Mikhael, and Ilan Mitnikov, the cross-disciplinary staff debuts their mannequin. The researchers present that ProtGPS can predict to which of 12 recognized kinds of compartments a protein will localize, in addition to whether or not a disease-associated mutation will change that localization. Moreover, the analysis staff developed a generative algorithm that may design novel proteins to localize to particular compartments.
“My hope is that it is a first step in direction of a strong platform that allows individuals finding out proteins to do their analysis,” Younger says, “and that it helps us perceive how people turn into the advanced organisms that they’re, how mutations disrupt these pure processes, and how one can generate therapeutic hypotheses and design medication to deal with dysfunction in a cell.”
The researchers additionally validated most of the mannequin’s predictions with experimental assessments in cells.
“It actually excited me to have the ability to go from computational design all the way in which to making an attempt this stuff within the lab,” Barzilay says. “There are a number of thrilling papers on this space of AI, however 99.9 p.c of these by no means get examined in actual techniques. Because of our collaboration with the Younger lab, we have been in a position to check, and actually learn the way properly our algorithm is doing.”
Growing the mannequin
The researchers skilled and examined ProtGPS on two batches of proteins with recognized localizations. They discovered that it might accurately predict the place proteins find yourself with excessive accuracy. The researchers additionally examined how properly ProtGPS might predict adjustments in protein localization primarily based on disease-associated mutations inside a protein. Many mutations — adjustments to the sequence for a gene and its corresponding protein — have been discovered to contribute to or trigger illness primarily based on affiliation research, however the methods wherein the mutations result in illness signs stay unknown.
Determining the mechanism for the way a mutation contributes to illness is essential as a result of then researchers can develop therapies to repair that mechanism, stopping or treating the illness. Younger and colleagues suspected that many disease-associated mutations would possibly contribute to illness by altering protein localization. For instance, a mutation might make a protein unable to hitch a compartment containing important companions.
They examined this speculation by feeding ProtGOS greater than 200,000 proteins with disease-associated mutations, after which asking it to each predict the place these mutated proteins would localize and measure how a lot its prediction modified for a given protein from the conventional to the mutated model. A big shift within the prediction signifies a probable change in localization.
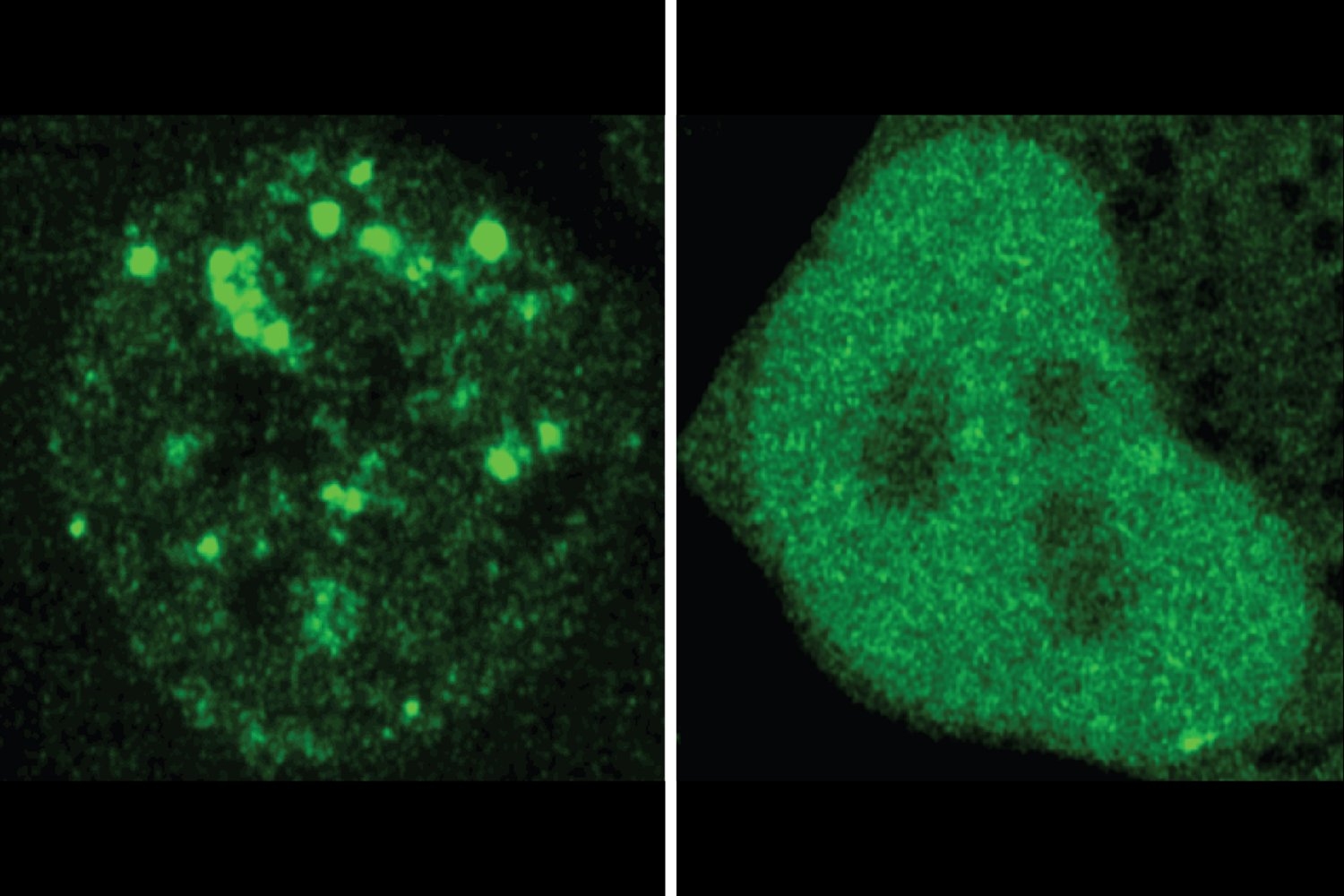
The researchers discovered many circumstances wherein a disease-associated mutation appeared to alter a protein’s localization. They examined 20 examples in cells, utilizing fluorescence to check the place within the cell a standard protein and the mutated model of it ended up. The experiments confirmed ProtGPS’s predictions. Altogether, the findings assist the researchers’ suspicion that mis-localization could also be an underappreciated mechanism of illness, and exhibit the worth of ProtGPS as a software for understanding illness and figuring out new therapeutic avenues.
“The cell is such an advanced system, with so many parts and sophisticated networks of interactions,” Mitnikov says. “It’s tremendous fascinating to suppose that with this method, we will perturb the system, see the result of that, and so drive discovery of mechanisms within the cell, and even develop therapeutics primarily based on that.”
The researchers hope that others start utilizing ProtGPS in the identical manner that they use predictive structural fashions like AlphaFold, advancing varied tasks on protein perform, dysfunction, and illness.
Shifting past prediction to novel technology
The researchers have been excited in regards to the attainable makes use of of their prediction mannequin, however in addition they wished their mannequin to transcend predicting localizations of present proteins, and permit them to design fully new proteins. The purpose was for the mannequin to make up completely new amino acid sequences that, when fashioned in a cell, would localize to a desired location. Producing a novel protein that may truly accomplish a perform — on this case, the perform of localizing to a selected mobile compartment — is extremely tough. So as to enhance their mannequin’s possibilities of success, the researchers constrained their algorithm to solely design proteins like these present in nature. That is an method generally utilized in drug design, for logical causes; nature has had billions of years to determine which protein sequences work properly and which don’t.
Due to the collaboration with the Younger lab, the machine studying staff was in a position to check whether or not their protein generator labored. The mannequin had good outcomes. In a single spherical, it generated 10 proteins supposed to localize to the nucleolus. When the researchers examined these proteins within the cell, they discovered that 4 of them strongly localized to the nucleolus, and others could have had slight biases towards that location as properly.
“The collaboration between our labs has been so generative for all of us,” Mikhael says. “We’ve realized how one can communicate one another’s languages, in our case realized lots about how cells work, and by having the prospect to experimentally check our mannequin, we’ve been ready to determine what we have to do to really make the mannequin work, after which make it work higher.”
Having the ability to generate useful proteins on this manner might enhance researchers’ skill to develop therapies. For instance, if a drug should work together with a goal that localizes inside a sure compartment, then researchers might use this mannequin to design a drug to additionally localize there. This could make the drug simpler and reduce unwanted side effects, because the drug will spend extra time participating with its goal and fewer time interacting with different molecules, inflicting off-target results.
The machine studying staff members are enthused in regards to the prospect of utilizing what they’ve realized from this collaboration to design novel proteins with different capabilities past localization, which might develop the probabilities for therapeutic design and different purposes.
“Plenty of papers present they’ll design a protein that may be expressed in a cell, however not that the protein has a specific perform,” Chinn says. “We truly had useful protein design, and a comparatively big success price in comparison with different generative fashions. That’s actually thrilling to us, and one thing we wish to construct on.”
All the researchers concerned see ProtGPS as an thrilling starting. They anticipate that their software shall be used to study extra in regards to the roles of localization in protein perform and mis-localization in illness. As well as, they’re excited by increasing the mannequin’s localization predictions to incorporate extra kinds of compartments, testing extra therapeutic hypotheses, and designing more and more useful proteins for therapies or different purposes.
“Now that we all know that this protein code for localization exists, and that machine studying fashions could make sense of that code and even create useful proteins utilizing its logic, that opens up the door for therefore many potential research and purposes,” Kilgore says.

