Detection of nucleotide mutations
Because of the restricted availability of lengthy RNA samples with outlined mutations, we first validated RNA-SCAN utilizing MS2 RNA as a mannequin service scaffold (Supplementary Fig. 2) and artificial nanolatches carrying particular base modifications (Fig. 2a). Particularly, we designed six nanolatch variants concentrating on the MS2 RNA web site, every carrying a definite sequence alteration: three with single-nucleotide substitutions at completely different positions (MHm1–MHm3), one with two substitutions (MHm4), one with an insertion (MHm5) and one with a deletion (MHm6). Every nanolatch was incubated individually with the service at a tenfold stoichiometric extra (Supplementary Fig. 3). The ensuing constructs had been analysed utilizing nanopore measurements.
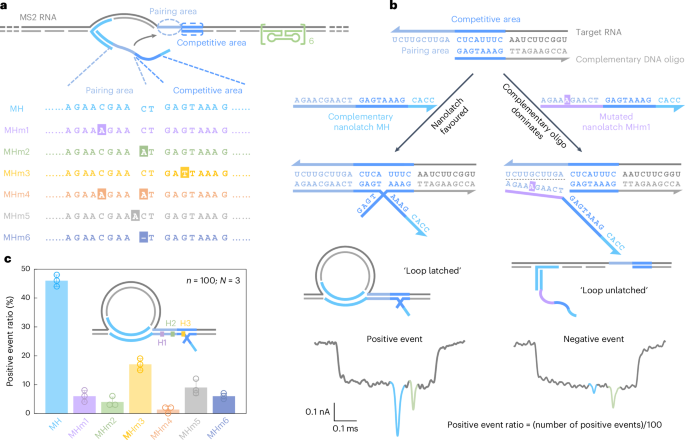
a, A sketch of the MS2 service design and sequence comparability between the complementary nanolatch (MH) and its six mutated variants (MHm1–MHm6), highlighting nucleotide modifications in each pairing and aggressive areas that work together with the goal web site on the MS2 service. b, The mechanism of the aggressive interplay between the MS2 RNA goal web site and nanolatches/complementary DNA oligo, illustrated utilizing the totally complementary nanolatch MH and single-nucleotide mutated nanolatch MHm1. The consultant nanopore present traces exhibit distinct signatures similar to the ‘loop latched’ (optimistic occasion) and ‘loop unlatched’ (adverse occasion) states. The optimistic occasion ratio is outlined because the variety of optimistic occasions divided by the entire variety of occasions per measurement (n = 100). c, An evaluation of optimistic occasion ratios for nanolatches MH–MHm6. The bar charts current optimistic occasion ratios from three unbiased measurements (N = 3) for every nanolatch. The info are proven because the imply ± commonplace deviation.
The sensitivity of RNA-SCAN to single-nucleotide variations stems from the finely tuned aggressive hybridization mechanism on the goal web site (Fig. 2b). Particularly, every goal web site is engineered with a 10-nt pairing space and an adjoining 8-nt aggressive space. When assembling the service, the aggressive space of the RNA scaffold is complemented by a DNA oligo, whereas the pairing space is deliberately left single-stranded to permit full nanolatch binding. This strategic design establishes a fragile competitors the place the goal RNA can transiently hybridize with both the nanolatch or the complementary DNA oligo. When the RNA sequence totally matches the nanolatch, their interplay is thermodynamically beneficial, yielding latched loop constructions, which manifest as attribute spike signatures in nanopore measurements (Fig. 2b, left). Nevertheless, even a single-nucleotide mismatch weakens the nanolatch-RNA binding affinity, tipping the stability in favour of the complementary oligo, which varieties a extra steady duplex with the RNA scaffold. This aggressive displacement of nanolatch will increase the probability of an unlatched state, characterised by the absence of the big present spike related to the latched configuration (Fig. 2b, proper). We classify translocation occasions as ‘optimistic’ when the attribute spike is current and ‘adverse’ when it’s absent. A present threshold of 0.55 (Prolonged Information Fig. 1) was utilized to facilitate classification. The optimistic occasion ratio is outlined because the variety of optimistic occasions out of 100 complete recorded occasions per nanopore measurement. Optimization of the pairing space and aggressive space is proven in Prolonged Information Fig. 3.
Determine 2c exhibits the optimistic occasion ratios calculated from the primary 100 linear nanopore occasions (n = 100) in every experiment, with error bars from three unbiased repeats (N = 3). This standardized quantification ensures dependable but environment friendly evaluation throughout circumstances (Supplementary Fig. 4). When the nanolatch sequence was totally complementary to the MS2 RNA goal web site (MH), the optimistic occasion ratio approached 50%. This stage displays the dynamic nature of loop formation and represents a stability optimized for single-nucleotide sensitivity. Within the presence of a single-nucleotide mismatch, the optimistic occasion ratio persistently dropped, although the magnitude of discount trusted the mutation’s place. Mutations within the pairing space (MHm1 and MHm2) led to an approximate tenfold lower, with optimistic occasion ratios falling to ~5%, whereas the change within the aggressive space (MHm3) resulted in the next common ratio of ~17%. That is in all probability as a result of mutations within the aggressive space primarily have an effect on the competitors between the nanolatch and the complementary oligo with out disrupting the preliminary nanolatch-RNA pairing, thereby permitting loop formation to proceed extra readily. To make sure excessive sensitivity and specificity of RNA-SCAN, we strategically positioned mutated or chemically modified nucleotides throughout the pairing space in subsequent designs. Two mismatches (MHm4) additional suppressed loop formation, virtually approaching the detection restrict. Nucleotide insertion (MHm5) or deletion (MHm6) within the nanolatch additionally disrupted its interplay with the service, lowering the optimistic occasion ratio to beneath 10%. These outcomes show that RNA-SCAN is extremely delicate to refined sequence variations and may discriminate single-base variations via simply interpretable nanopore signatures (Prolonged Information Fig. 4).
Detection of nucleotide modifications
Having demonstrated the flexibility of RNA-SCAN to detect single-nucleotide mutations, we subsequent utilized this system to epigenetic nucleotide modifications, which play a vital function in lots of elementary metabolic features3,4.
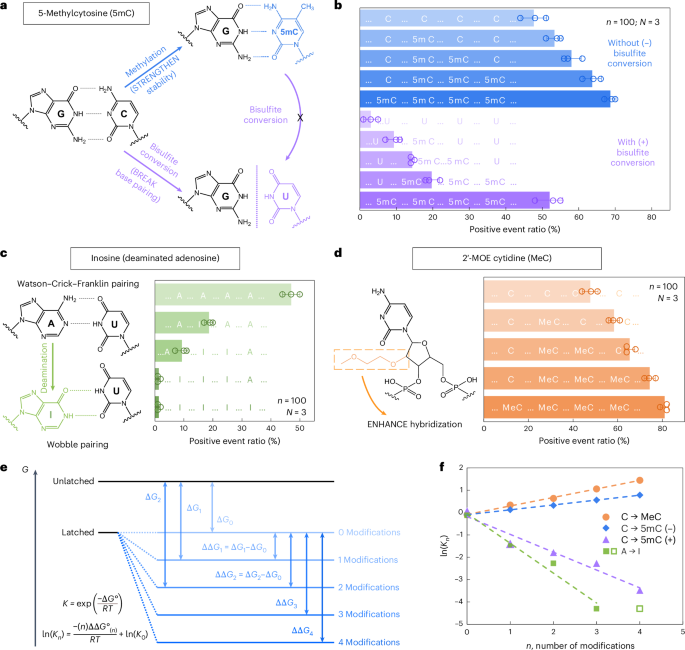
We first examined 5-methylcytosine (denoted as 5mC in DNA and m5C in RNA), a key epigenetic marker incessantly noticed in most cancers21,22. To research its presence and abundance in nucleic acids, we evaluated the RNA-SCAN system from two complementary instructions. First, the covalent introduction of a methyl group to cytosine alters the van der Waals radii across the C5 place and modifies base stacking potential. Prior research have proven that such methylations can improve duplex stability by influencing thermodynamic properties23,24. Alternatively, bisulfite therapy can selectively deaminate unmethylated cytosines to uracil whereas leaving 5-methylcytosines intact25,26. This chemical conversion alters the base-pairing desire of cytosine from guanine to adenine, thereby disrupting hybridization patterns (Fig. 3a). Following these ideas, we designed 5 nanolatch variants containing 0–4 5mC modifications and measured loop formation with and with out bisulfite conversion. The nanopore measurement outcomes are summarized in Fig. 3b. With out bisulfite conversion, rising the variety of 5mC modifications led to greater optimistic occasion ratios, reaching ~69% for the four-modification nanolatch, which is sort of 1.5 occasions the ratio noticed with the unmodified management (~48%). With bisulfite conversion, the optimistic occasion ratio for the four-methylated nanolatch remained comparatively excessive at ~52%, whereas nanolatches containing fewer 5mC modifications exhibited a considerable lower in optimistic occasions. This sample means that bisulfite-induced deamination of unmethylated cytosines disrupted their pairing with guanines, destabilizing loop formation and resulting in unlatching.
a, A schematic depicting how 5mC modification influences C/G base pairing earlier than and after bisulfite conversion. The MS2 service design is equivalent to that in Fig. 2. b, The nanopore measurement outcomes of MS2 carriers incubated with nanolatches containing 0–4 5mC modifications, with (+) and with out (−) bisulfite therapy. c, A schematic of how inosine modification impacts A/U base pairing, together with nanopore measurement outcomes for nanolatches containing 0–4 inosines within the sequence. d, A schematic of MeC modification and nanopore measurement outcomes for nanolatches possessing 0–4 MeCs. The info in b–d are proven because the imply ± commonplace deviation from three unbiased measurements. e, A theoretical mannequin of the nanolatch system for detecting modifications. The presence of modifications can both strengthen or weaken the steadiness of the duplex between the nanolatch and the RNA scaffold, altering the power distinction (Delta G) between ‘latched’ and ‘unlatched’ power ranges. R is the perfect gasoline fixed, T is absolutely the temperature at which the response happens, ({Ok}_{n}) is the equilibrium fixed of loop latching, and (n) is the variety of modifications. f, The linear becoming outcomes of (mathrm{ln}({Ok}_{n})) in opposition to (n) for every kind of modifications studied on this work. 5mC (+) and 5mC (−) check with cytosine methylations with (+) and with out (−) bisulfite therapy, respectively. The info level for 4 inosines (open sq.) was excluded from becoming, because the optimistic occasion ratio had already dropped to just about 0% with three inosines.
We subsequent examined the flexibility of RNA-SCAN to detect inosine, a product of adenosine deamination that’s related to RNA modifying and post-transcriptional gene regulation27,28. Chemically, this amino-to-keto conversion alters the bottom’s hydrogen bonding properties, remodeling adenosine from a hydrogen bond donor into an acceptor. Consequently, canonical Watson–Crick–Franklin base pairing with uridine is changed by two weaker, non-canonical wobble hydrogen bonds29,30 (Fig. 3c, left). This transformation destabilizes the RNA/DNA duplex and alters loop formation throughout the service. To guage this impact, we designed 5 nanolatches containing 0–4 inosine substitutions. Nanopore measurements (Fig. 3c, proper) confirmed that even a single inosine substitution decreased the optimistic occasion ratio from ~47% to ~19%. With three inosines current, loop formation was virtually utterly disrupted, and the sign dropped to close zero. These outcomes verify that RNA-SCAN can sensitively detect the structural penalties of inosine incorporation.
To evaluate RNA-SCAN’s capability to detect sugar modifications, we integrated 2′-O-methoxyethyl (2′-MOE) teams into cytidines (2′-MOE cytidine, MeC), that are recognized to boost duplex stability in antisense oligo therapeutics31,32 (Fig. 3d, left). Substituting 5mC with MeC in nanolatches led to even better optimistic indicators (Fig. 3d, proper): 4 MeC modifications yielded ~81% optimistic occasions, in contrast with ~69% with 5mC and ~48% with unmodified sequences. Even a single MeC elevated sign by over 10%, highlighting this modification’s robust stabilizing impact on base pairing.
To interpret these findings, we modelled loop formation as a two-state power system: latched versus unlatched (Fig. 3e). Modifications shift the power of the latched state, altering its equilibrium. Primarily based on the nearest-neighbour mannequin33,34, which assumes additive energetic contributions from adjoining base pairs, we noticed robust linear correlations between the variety of modifications and the equilibrium fixed (Fig. 3f). These developments aligned with our experimental outcomes and assist the thermodynamic foundation for modification detection. Additional particulars can be found in Supplementary Observe 2.
Discrimination of E. coli and S. Typhi by RNA-SCAN
To guage the efficiency of RNA-SCAN on naturally occurring lengthy RNA, we utilized it to distinguish between Escherichia coli and Salmonella enterica serovar Typhi (S. Typhi), two carefully associated Gram-negative micro organism of medical relevance. Whereas most E. coli strains are innocent intestine commensals, some pathogenic variants trigger extreme gastrointestinal sickness35,36. S. Typhi, a human-specific pathogen, is accountable for enteric (typhoid) fever and presents distinct antimicrobial resistance profiles in contrast with E. coli37,38.
To allow species-level discrimination, we centered on 16S ribosomal RNA (rRNA), a extremely conserved however diagnostically informative bacterial id marker. Sequence alignment throughout all 14 operons from each species revealed >99% intraspecies similarity and 97–98% interspecies similarity (Supplementary Fig. 5). Inside this context, we recognized an 18-nt area containing a constant single-nucleotide distinction between E. coli and S. Typhi 16S rRNA and chosen it because the goal for RNA-SCAN detection (Fig. 4a,b). Separate oligo swimming pools had been designed to assemble RNA carriers from the entire 16S rRNA of every species, whereas nanolatches had been tailor-made to match the species-specific variant of the goal sequence. Every service was incubated with both a matched or mismatched nanolatch, creating 4 experimental combos.
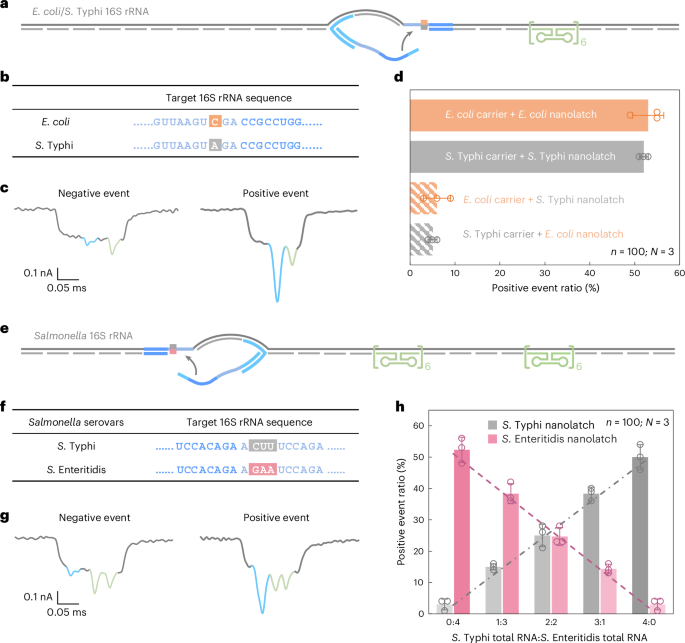
a, A schematic of the E. coli/S. Typhi 16S rRNA service design with a reference construction positioned on one aspect of the service and a goal web site situated on the centre. b, The sequences of E. coli and S. Typhi 16S rRNA on the goal web site, highlighting a single-nucleotide variation. c, Consultant nanopore present traces of the E. coli/S. Typhi 16S rRNA service. d, The evaluation of the optimistic occasion ratio for the 4 combos of E. coli/S. Typhi service with E. coli/S. Typhi nanolatch. The service focus was set at 0.3 nM firstly of nanopore measurements and the nanolatch focus at 3 nM for each E. coli and S. Typhi. e, A schematic of the Salmonella 16S rRNA service design with two reference constructions positioned on one aspect and a goal web site on the opposite aspect. f, The sequences of S. Typhi and S. Enteritidis 16S rRNA on the goal web site, highlighting three steady completely different bases. g, Consultant nanopore present traces of the Salmonella 16S rRNA service. h, An evaluation of the optimistic occasion ratio for complete RNA mixtures of S. Typhi and S. Enteritidis at outlined enter ratios, utilizing both the S. Typhi or S. Enteritidis nanolatch. The mixed focus of S. Typhi and S. Enteritidis 16S rRNA carriers was maintained at 0.3 nM in all nanopore measurements, with a relentless nanolatch focus of three nM. The info in d and h are proven as imply ± commonplace deviation from three unbiased measurements.
Nanopore measurements had been carried out utilizing complete RNA immediately extracted from E. coli and S. Typhi cultures, with out extra purification or enrichment. In matched combos—E. coli service with E. coli nanolatch or S. Typhi service with S. Typhi nanolatch—the optimistic occasion ratio exceeded 50%, indicating profitable loop formation (Fig. 4d). Against this, the mismatched combos yielded optimistic occasion ratios of round 5%, demonstrating a transparent lack of nanolatch binding. This almost tenfold distinction in sign underscores the excessive sensitivity and specificity of RNA-SCAN in distinguishing single-nucleotide variations inside lengthy, native RNA molecules (Prolonged Information Fig. 5). Importantly, the assay requires no enzymatic processing, amplification or fragmentation and works immediately with complete bacterial RNA, making it extremely appropriate for sensible functions equivalent to microbial identification and resistance profiling.
Quantification of Salmonella 16S rRNA variants
Whereas the excessive sequence similarity of 16S rRNA between E. coli and S. Typhi already poses a problem for standard assays, completely different serovars of Salmonella enterica, equivalent to S. Typhi and S. Enteritidis39, are much more tough to differentiate, with 16S rRNA sequence identities exceeding 99.5% overlap (Supplementary Fig. 6).
To deal with this, we designed a shared oligo pool that binds conserved areas in each S. Typhi and S. Enteritidis 16S rRNA, utilizing consensus nucleotides at polymorphic websites. The service design included two reference constructions on one aspect to differentiate this assemble from earlier E. coli/S. Typhi carriers (Fig. 4e and Prolonged Information Fig. 6) and a nanolatch binding web site on the opposite aspect the place three consecutive nucleotides differ between the 2 serovars (Fig. 4f). To imitate medical circumstances the place RNA from a number of bacterial species might coexist, we combined complete RNA extracted from S. Typhi and S. Enteritidis in outlined ratios. After hybridizing the combination with the oligo pool to generate carriers for each serovars, every combination was cut up and incubated with both a S. Typhi-specific or S. Enteritidis-specific nanolatch.
The nanopore measurements confirmed that the optimistic occasion ratio elevated linearly with the proportion of every goal RNA within the combination when probed with its matching nanolatch (Fig. 4h). This linear relationship confirms that RNA-SCAN can’t solely distinguish between extremely comparable sequences but in addition quantify their relative abundance in complicated RNA backgrounds. The small error bars and constant matches additional point out that different minor sequence variations or intraspecies polymorphisms didn’t intrude with detection. These outcomes spotlight the potential of RNA-SCAN for multiplexed pathogen quantification in combined samples, with single-nucleotide decision and with out requiring prior pattern purification or amplification.
m5C detection in 16S rRNA from E. coli and A. baumannii
Past sequence variation, we additional evaluated the flexibility of RNA-SCAN capability to detect base modifications in endogenous lengthy RNA. In E. coli, rRNAs comprise at the least 24 recognized methylation websites important for ribosomal operate40. Amongst these, three are m5C modifications situated at positions C967 and C1407 in 16S rRNA (Fig. 5a) and at C1962 in 23S rRNA. Notably, methylation at C1407 is extremely conserved in E. coli and has been implicated in translation constancy40,41. Against this, Acinetobacter baumannii, a standard reason for healthcare-associated opportunistic infections, utterly lacks the methyltransferase gene accountable for C1407 methylation42,43. This makes E. coli and A. baumannii perfect comparative fashions for finding out RNA methylation at this place.
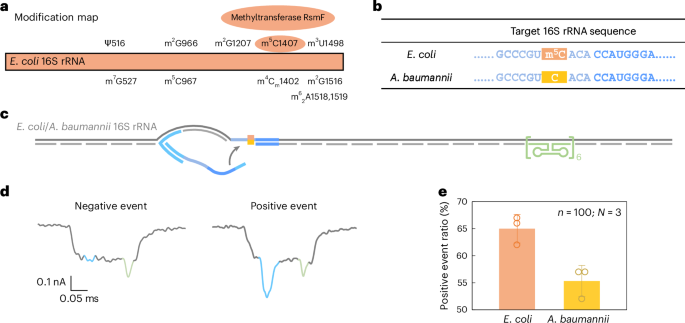
a, A modification map of E. coli 16S rRNA, highlighting the important function of the methyltransferase RsmF in catalysing methylation at cytosine 1407. b, The sequences of E. coli and A. baumannii 16S rRNA on the goal web site. The cytosine at place 1407 is methylated in E. coli 16S rRNA however stays unmethylated in A. baumannii 16S rRNA. c, A schematic of the E. coli/A. baumannii 16S rRNA service design, that includes a reference construction on one aspect of the service and the goal web site on the reverse finish. d, Consultant nanopore present traces of the E. coli/A. baumannii 16S rRNA service. e, The evaluation of the optimistic occasion ratio for E. coli and A. baumannii 16S rRNA carriers utilizing the identical nanolatch. The service focus was maintained at 0.3 nM and the nanolatch focus at 3 nM. The upper optimistic occasion ratio noticed for E. coli will be attributed to the stabilizing impact of the m5C modification on the goal web site. The info are proven as imply ± commonplace deviation from three unbiased measurements.
Given the substantial sequence divergence of over 15% between their 16S rRNA operons (Supplementary Fig. 7), we designed species-specific oligo swimming pools for assembling RNA carriers, whereas utilizing a standard nanolatch concentrating on the C1407 area. The ensuing service construction included the C1407 goal web site on one aspect and a reference construction on the opposite (Fig. 5b,c). The full RNA extracted immediately from bacterial cells was used with out additional processing.
Nanopore measurements confirmed a statistically vital distinction in loop formation between species. E. coli produced a optimistic occasion ratio of 65% ± 2%, in contrast with 55% ± 2% for A. baumannii (Fig. 5e and Prolonged Information Fig. 7), with a t-test yielding P = 0.01289. These outcomes are in step with our earlier MS2-based experiments, supporting the interpretation that m5C1407 enhances nanolatch binding by stabilizing C/G base pairing. Apparently, each species exhibited greater sign ranges than unmodified MS2 carriers. To research this, we carried out bisulfite sequencing on the 16S rRNA goal area. The outcomes (Prolonged Information Fig. 8) counsel that one other close by modification, m4Cm1402 (a 2′-O-methylated and N4-methylated cytosine present in each species), is partly proof against bisulfite conversion and will equally strengthen base pairing.
Collectively, these findings show the potential of RNA-SCAN to detect single-base RNA modifications in native, unamplified samples. Furthermore, the commentary of enhanced indicators from close by modified nucleotides means that RNA-SCAN might additionally function a software for figuring out beforehand unannotated modifications in functionally necessary RNA areas.